Protocol
EpiMelt Bisulfite Modification Standard Kit
Complete kit for efficient conversion and preparation of converted DNA for methylation research.

For 50 & 200 reactions
| Catalog # | Size |
| # EPI – BIS – 50 | 50 reactions |
| # EPI – BIS – 200 | 200 reactions |
EpiMelt Bisulfite Modification Standard Kit
Highlights
- Streamlined proven procedure for bisulfite conversion of DNA.
- Desulphonation and recovery of bisulfite-treated DNA using a spin column.
- The recovered DNA is ideal for downstream analyses including PCR, qPCR followed by melting
analysis (MS-HRM, qMSP), sequencing, microarray analyses, etc.
Introduction to DNA Methylation
DNA methylation and histone modifications are naturally occurring epigenetic mechanisms, which can modulate gene expression in response to the environment.
DNA Methylation is a covalent addition of a methyl group to the carbon-5 position of cytosine residues within 5’-CpG-3’ dinucleotides. These CpG sites are not evenly distributed throughout the genome but are clustered in CpG-rich regions, known as CpG islands (CGI). A CpG island is typically between 500-1000 base pairs (bp) long and spans regulatory regions of the genome for example gene promoter regions. More than half of the protein-coding genes contain a CpG island spanning their promoter region, in which the methylated status is instrumental in regulating the transcriptional activity of the gene.
To detect and quantify DNA methylation accurately and efficiently has become highly essential in the study of cancer, as well as for example in age and lifestyle-related diseases, gene-expression studies, and genetic diseases. In cancer research, the DNA methylation profile can be associated with a predisposition, diagnosis, prognosis, and prediction of treatment response, and allows to following residual disease. An example of aberrant DNA methylation pointing to predisposition to disease is that promoter methylation of the BRCA1 gene detected in peripheral blood is associated with the risk of developing triple-negative breast cancer and high-grade serous ovarian cancer. DNA methylation assays targeting methylation of the MLH1 gene are used in the pre-screening and diagnosis of Lynch syndrome. The methylation status of the MGMT gene has for decades been used to triage glioblastoma patients for temozolomide treatment. Determining the methylation status of the BRCA1 gene has been argued to be included in the selection of patients for the PARP inhibitor treatment. Finally, assessment of the methylation status of C9orf50, CLIP4, and KCNQ5 promoter regions detected in peripheral blood improves colorectal cancer relapse detection after surgery.
Different techniques are developed to measure DNA methylation levels at specific genomic sites, and with few exceptions, all are dependent on bisulfite conversion of the DNA prior to applying the DNA methylation assessment method.
During bisulfite treatment, the unmethylated cytosines are converted into uracils, whereas methylated cytosines are protected from conversion and remain unchanged (Figure 1). The DNA sequence of the methylated and unmethylated DNA strands are therefore different after bisulfite conversion.
Table 1. Contents of the EpiMelt Real-Time PCR Master Mix.
Template DNA:
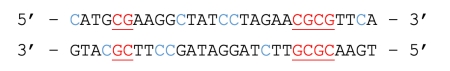
Bisulfite converted DNA:
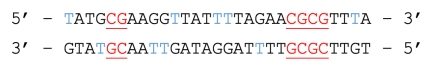
Figure 1. The underlined and red bases represent CpG dinucleotides containing a methylated cytosine, which is preserved during bisulfite treatment. The blue bases represent unmethylated cytosines, converted to uracil during bisulfite treatment and replaced by thymine during PCR.
Methylation-Sensitive High-Resolution Melting (MS-HRM) is an example of a technology for post-bisulfite modification detection of DNA methylation. MS-HRM enables cost-effective, fast, and highly sensitive assessment of locus-specific methylation status.
The principle of MS-HRM is that sequence-specific melting profiles of amplicons are observed across a gradually increasing temperature gradient in the presence of an intercalating, fluorescent dye. As the targets are amplified from bisulfite-converted template DNA, the amplicons from the methylated template DNA will have a high GC content, and amplicons from the unmethylated template will have a low GC content. The amplification is performed using a unique primer design developed to overcome PCR bias. Amplicons from the methylated template will then melt at a higher temperature than amplicons from the unmethylated template. This principle allows the assessment of methylation status based on the amplicon melting profile (Figure 2). The inclusion of methylation positive and negative controls allows for a semi-quantitative estimation of the methylation status of a test sample.
For research use only. Not for use in diagnostic procedures.
A
Un-methylated sequence
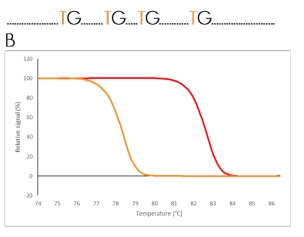
A
Methylated sequence
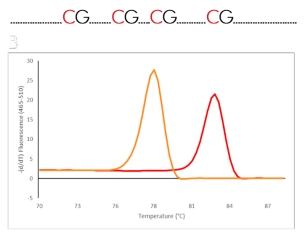
Figure 2. Schematic illustration of the principle behind High-Resolution Melting (HRM) analysis after qPCR of a bisulfite converted
template. A) The difference in a DNA sequence after bisulfite conversion of an unmethylated and a methylated region. B) The
difference in melting properties of the PCR products from the methylated (red) and the unmethylated (orange) templates
displayed as melting curves and melting peaks.
Download the Protocol here:
Further reading
Clark, S. J., Statham, A., Stirzaker, C., Molloy, P. L. & Frommer, M. DNA methylation: Bisulphite modification and
analysis. Nature protocols 1, 2353-2364 (2006). https://doi.org:10.1038/nprot.2006.324
Nishiyama, A. & Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet 37, 1012-
1027 (2021). https://doi.org:10.1016/j.tig.2021.05.002
Mattei, A. L., Bailly, N. & Meissner, A. DNA methylation: a historical perspective. Trends Genet 38, 676-707 (2022).
https://doi.org:10.1016/j.tig.2022.03.010
Wojdacz TK, Dobrovic A, Hansen LL: Methylation-sensitive high-resolution melting. Nature protocols 2008,
3(12):1903-1908 https://doi:10.1038/nprot.2008.191
Specifications:
Starting materials:
DNA: purified from tissue culture, solid tissue, formalin-fixed paraffin-embedded (FFPE) tissue, whole blood, buffy coat, biopsies, laser-captured micro-dissected (LCM) tissue, etc.
DNA Input: Samples containing 500 pg – 2 μg of DNA. For optimal results, the recommended amount of input DNA should be from 200 – 500 ng.
For research use only. Not for use in diagnostic procedures.
Product Description
The EpiMelt Bisulfite Modification Standard Kit features a simple, column-based procedure for streamlined, high-throughput bisulfite conversion of DNA. The kit includes a set of reagents for bisulfite conversion and purification of converted DNA that has been designed to minimize template degradation and loss of DNA during treatment and clean-up as well as to provide a high rate of conversion of unmethylated cytosines. Purified, converted DNA is suited for all downstream techniques currently used for the analysis of DNA methylation, including qPCR-based technologies followed by high-resolution melting analysis (MS-HRM), pyrosequencing, and bisulfite sequencing.
The yield of DNA after bisulfite conversion and clean-up is dependent on the amount and quality of the sample DNA used for conversion. The optimal results obtained by the EpiMelt Bisulfite Modification Standard Kit are based on DNA input ranging from 500 pg to 2 μg. The cytosine conversion rate leads to equal DNA recoveries after purification of the converted DNA independent of DNA starting input.
Kit Contents
| Component | Quantity | Quantity | Storage |
| Microcolumns with 2 mL tubes | 50 pcs | 200 pcs | Room temperature |
| C/T Conversion Reagent | 5 tubes | 20 tubes | Room temperature |
| D Dilution Solution | 1.5 mL | 5 mL | Room temperature |
| G Binding Solution | 35 mL | 130 mL | Room temperature |
| A1 Wash Solution | 30 mL | 110 mL | Room temperature |
| DS Desulphonation Solution | 12 mL | 45 mL | Room temperature |
| Tris Buffer | 2 mL | 8 mL | Room temperature |
| Ultrapure Water | 8 mL | 30 mL | Room temperature |
For research use only. Not for use in diagnostic procedures.
Intended use
The EpiMelt Bisulfite Modification Standard Kit is intended for research applications only. This product is neither intended for diagnosis, prevention, or treatment of a disease, nor has it been validated for such use, either alone or in combination with other products.
For safety information, please refer to page 9 of this protocol or visit www.MethylDetect.com.
Additional Equipment and Reagents
- 1.5 mL sterile microtubes
- Vortex
- Microcentrifuge
- Thermoblock
Important Notes
- Before use, all reagents should be gently mixed by inverting the tubes and briefly centrifuged.
- The C/T Conversion Reagent is supplied as a solid crystalline in an amber tube and is light sensitive.
For best results, C/T Conversion Reagent should be used immediately after solution preparation.
For research use only. Not for use in diagnostic procedures.
Download the Protocol here:
Protocol
Reagent Preparation
Preparation of C/T Conversion Reagent
- Add 750 μL Ultrapure Water and 210 μL D Dilution Solution to the C/T Conversion Reagent tube.
- Mix by vortexing or shaking for 10 minutes.
Note: Each tube of C/T Conversion Reagent is designed for 10 separate DNA treatments.
Note: The C/T Conversion Reagent can be stored overnight at room temperature, up to one week at 4°C, or up to one month at -20°C
DNA Conversion Protocol
The conversion reaction can process a sample containing 500 pg to 2 μg of DNA. For optimal results, the amount of input DNA should be within a range of 200 – 500 ng.
- Add Ultrapure Water to DNA samples up to a total volume of 50 μl.
Add 100 μL C/T Conversion Reagent to each sample and mix by pipetting.
Important: Do not vortex - Incubate the samples in the dark for 10 min at 98°C and then 2.5 h at 64°C
- Cool down samples for 10 min on ice (0-4°C)
Note: At this point, the samples can be stored at 4°C for up to 20 h.
Purification of Converted DNA
- Add 600 μL G Binding Solution to each tube. Mix by inverting the tubes.
- Transfer the mixtures into the microcolumns. Close the tubes with the caps.
- Centrifuge for 30 – 60 s at ≥ 10 000 g (10 000 – 15 000 rpm).
- Remove the microcolumns from the tubes and discard flow-through. Place the microcolumns back into the same tubes.
- Apply 100 μL A1 Wash Solution to the microcolumns. Close the tubes with the caps.
- Centrifuge for 30 – 60 s at ≥ 10 000 g (10 000 – 15 000 rpm).
- Apply 200 μL DS Desulphonation Solution to the microcolumns. Close the tubes with the caps. Incubate for 10 min at room temperature.
- Centrifuge for 30 – 60 s at ≥ 10 000 g (10 000 – 15 000 rpm).
- Remove the microcolumns from the tubes and discard flow-through. Place the microcolumns back into the same tubes.
- Apply 200 μL A1 Wash Solution to the microcolumns. Close the tubes with the caps.
- Centrifuge for 30 – 60 s at ≥ 10 000 g (10 000 – 15 000 rpm).
- Apply 200 μL A1 Wash Solution to the microcolumns. Close the tubes with the caps.
- Centrifuge for 2 min s at ≥ 10 000 g (10 000 – 15 000 rpm).
- Transfer dried microcolumns into sterile 1.5 mL microtubes for elution (microtubes are not included).
Add 15 – 30 μL Tris Buffer or Ultrapure Water directly to the microcolumn resin.
Attention: When adding elution buffer to the microcolumns, ensure that the liquid is applied precisely onto the resin. If liquid stays on the column walls, the elution may not be effective. - Incubate the microcolumns for 3 min at room temperature.
- Centrifuge for 1 min at ≥ 10 000 g (10 000 – 15 000 rpm).
- Remove microcolumns and close the elution tubes.
Note: Store the purified DNA samples at 4°C to 8°C
Frequently Asked Questions
Q: Should the input DNA be dissolved in TE or water?
A: The input DNA can be dissolved in TE or water, as neither of these will interfere with the conversion process.
Q: Do you recommend a specific Taq polymerase for PCR of the bisulfite converted DNA?
A: A hot start Taq polymerase is recommended (e.g., the EpiMelt Real-Time PCR Master Mix).
For research use only. Not for use in diagnostic procedures.
Safety Information

A1 Wash Solution
H255 Highly flammable liquid and vapor.
H319 Causes serious eye irritation.
H336 May cause drowsiness or dizziness.
P210 Keep away from heat, sparks, open flames, hot surfaces. No smoking.
P261 Avoid breathing vapors.
P305+P351+P338 If in eyes: Rinse cautiously with water for several minutes.
Remove contact lenses, if present and easy to do. Continue rinsing

C/T Conversion Reagent
H302 Harmful if swallowed.
H319 Causes eye irritation.
P264 Wash skin thoroughly after handling.
P270 Do not eat, drink, or smoke when using this product.
P280 Wear protective gloves/protective clothing/eye protection/face protection.
P301+P312 If swallowed: Call a Poison Center/doctor if you feel unwell.
P305+P351+P338 If in eyes: Rinse cautiously with water for several minutes.
Remove contact lenses, if present and easy to do. Continue rinsing.
P337+P313 If eye irritation persists: Get medical advice/attention

D Dilution Solution
H290 Corrosive to metals.
H314 Causes eye irritation.
P260 Do not breathe dust.
P280 Wear protective gloves/protective clothing/eye protection/face protection.
P303+P361+P353 If on skin (or hair): Take off all contaminated clothing
immediately. Rinse skin with water.
P304+P340+P310 If inhaled: Remove person to fresh air and keep comfortable
for breathing. Immediately call a Poison center/doctor.
P305+P351+P338 If in eyes: Rinse cautiously with water for several minutes.
Remove contact lenses, if present and easy to do. Continue rinsing.
P310 Immediately call a Poison Center/doctor

DS Desulphonation Solution
H255 Highly flammable liquid and vapor.
H290 Corrosive to metals.
H314+H319 Causes serious eye and skin irritation.
H336 May cause drowsiness or dizziness.
P210 Keep away from heat, sparks, open flames, hot surfaces. No smoking.
P261 Avoid breathing vapors.
P280 Wear protective gloves/protective clothing/eye protection/face protection.
P305+P351+P338 If in eyes: Rinse cautiously with water for several minutes.
Remove contact lenses, if present and easy to do. Continue rinsing.
P310 Immediately call a Poison Center/doctor

G Binding Solution
H302 Harmful if swallowed.
H315 Causes skin irritation.
H319 Causes serious eye irritation.
P305+P351+P338 If in eyes: Rinse cautiously with water for several minutes.
Remove contact lenses, if present and easy to do. Continue rinsing
Ordering Information
MethylDetect offers EpiMelt Methylation Detection Assays targeting specific genomic regions. For a complete overview of the products and ordering information, please visit www.MethylDetect.com.
Supplementary Information
License Disclaimer
For patent license limitations for individual products, please refer to www.MethylDetect.com.
Regulatory Disclaimer
For Life Science research only. Not for use in diagnostic procedures.
Safety Data Sheet
Please follow the instructions in the safety data sheet (SDS) at www.MethylDetect.com.
Contact and Support
Please refer to www.MethylDetect.com.
For research use only. Not for use in diagnostic procedures.
©2023 MethylDetect. All rights reserved.
