APPLICATION NOTE
Bisulfite Modification Standard Kit
High bisulfite conversion efficiency using EpiMelt Bisulfite Modification Standard Kit on genomic DNA
Ea Marie Givskov Sørensen, Tina Ellegaard Kjeldsen, Lise Lotte Hansen
MethylDetect ApS, Aalborg, Denmark

Abstract
DNA methylation biomarkers are becoming increasingly relevant in assessing disease predisposition, diagnosis and detection of early-stage disease, personalized treatment, and post-treatment surveillance. In research, methylation analyses are often performed on limited or degraded biological material. Therefore, the sensitivity of the detection method is essential.
EpiMelt Methylation Detection Assays are based on the Methylation-Sensitive High Resolution Melting (MS-HRM) technology, where the unique primer design and specific annealing temperature of the assay ensure robust and highly sensitive DNA methylation analysis.
In this application note, we show the performance of five EpiMelt Methylation Detection Assays and the EpiMelt Bisulfite Modification Standard Kit.
We demonstrate this to be an efficient and sensitive method of bisulfite modification and methylation detection.
Introduction
DNA methylation is an epigenetic modification, which primarily consists of a methyl moiety attached to cytosines in CpG dinucleotides. In healthy cells, CpGs in gene-coding and non-gene-coding regions are generally methylated. At the same time, first exons, promoters, and enhancers containing high levels of CpG-content (CpG islands) typically are unmethylated1,2. Hypomethylation is often a sign of active transcription, while hypermethylation is associated with gene transcription silencing3. However, the established DNA methylation patterns are disrupted in cancer, and CpG island methylation status at specific genes can have distinct consequences for patients at all stages of the disease.
One of the most common methods of investigating DNA methylation status is bisulfite conversion followed by a method of methylation detection. The bisulfite treatment converts unmethylated cytosines into uracils, while methylated cytosines are protected1. Bisulfite conversion involves the use of harsh chemicals, which induces degradation of the DNA that then risks being lost during the subsequent DNA purification4-6. However, the EpiMelt Bisulfite modification Standard Kit employs a bisulfite conversion step with high temperature and bisulfite concentration, and decreased processing time, a technique that efficiently and homogenously converts DNA6,7.
The EpiMelt Methylation Detection Assays are based on Methylation-Sensitive High Resolution Melting (MS-HRM). MS-HRM is the methylation-independent amplification of the target region, followed by HRM8. During HRM, the fluorescence signal from a dsDNA intercalating dye is measured across an increasing temperature gradient. The melting profiles of the amplicons reflect whether the template DNA was methylated, as it will have a high GC content and therefore melts at a higher temperature, compared to amplicons from unmethylated template DNA9. The primers are designed to match the methylated
template, to overcome PCR bias8. The EpiMelt Methylation Detection Assays contain a locus-specific primer mix and three ready-to-use controls (Methylation Positive Control, Assay Calibration Control, and Methylation Negative Control). The method is semiquantitative, and the methylation status of an unknown sample can be determined by comparison to the melting profiles of the three controls10.
In this application note, we demonstrate the robust and efficient bisulfite conversion and clean-up of formalin-fixed paraffin-embedded (FFPE) non-small-cell lung cancer samples, and the sensitive methylation detection using the EpiMelt Bisulfite modification Standard Kit in combination with the EpiMelt methylation detection assays targeting BRCA1, BRCA2, HORMAD1, MGMT1, and MLH1.
Materials
Reagents and consumables
Genomic DNA (gDNA) DNA was extracted from 5 x 10 μm from non-small -cell lung cancer (NSCLC) formalin-fixed paraffin-embedded (FFPE) sections using the High Pure FFPET DNA Isolation Kit (Roche, Germany) according to the manufacturer’s protocol and stored in TE buffer. Samples were divided into low, medium, and high-quality DNA based on an agarose gel electrophoresis.
Materials
Qubit™ 1 x dsDNA HS Assay Kit
(Thermo Fisher Scientific, cat. #Q33230).
EpiMelt Bisulfite Modification Standard Kit
(MethylDetect ApS, cat. # EPI-BIS-200)
EpiMelt BRCA1 test assay (MethylDetect ApS)
EpiMelt BRCA2 test assay (MethylDetect ApS).
EpiMelt HORMAD1test assay (MethylDetect ApS).
EpiMelt MGMT test assay (MethylDetect ApS).
EpiMelt MLH1 test assay (MethylDetect ApS).
EpiMelt Real-Time PCR Master Mix
(MethylDetect ApS, cat. # EPI-qPCR-200).
LightCycler® 480 Multiwell plate 96, white
(Roche, cat. # 04729692001).
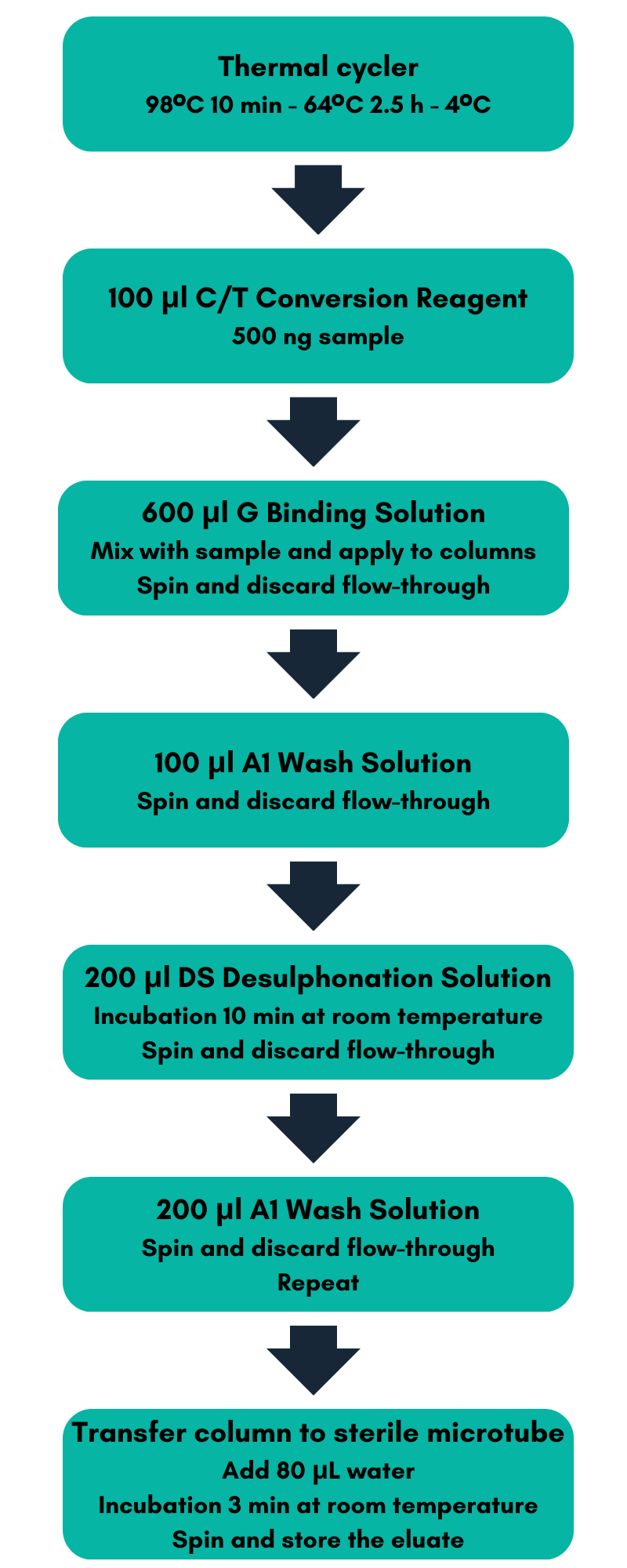
Figure 1:
Bisulfite conversion and purification workflow.
Download the Application Note here:
Instruments
Qubit™ 3 Fluorometer (Thermo Fisher Scientific, cat. # Q33216).
LightCycler® 480 System (Roche, cat. # 05015278001).
Methods
Bisulfite conversion and purification
Prior to bisulfite conversion and DNA purification, the gDNA concentration was measured using the Qubit™ 1 x dsDNA HS Assay on the Qubit™ 3 Fluorometer.
Water and D Dilution Buffer were added to the C/T Conversion Reagent from the EpiMelt Bisulfite Modification Standard Kit to solubilize the solid crystalline. For bisulfite conversion, 500 ng gDNA from each of the three samples was diluted with water to a volume of 50 μL and used as input. Bisulfite treatment and DNA purification were performed using the EpiMelt Bisulfite Modification Standard Kit according to the workflow depicted in Figure 1.
Amplification and high-resolution melting
Five EpiMelt Methylation Detection Assays were used to analyze the gDNA samples, targeting BRCA1, BRCA2, HORMAD1, MGMT, and MLH1. Each EpiMelt Methylation Detection Assay contains a locus-specific primer mix and three ready-to-use controls. The EpiMelt Controls or the converted and purified gDNA samples were combined with 10.0 μL EpiMelt Real-Time PCR Master Mix, 3.0 μL water, and 1.0 µL EpiMelt Primer Mix according to Table 1.
| Reagent | Volume |
| EpiMelt Real-Time PCR Master Mix 2 x | 10.0 μL |
| Nuclease-free water | 3.0 μL |
| EpiMelt Primer Mix (10 mM) | 1.0 μL |
| EpiMelt Control (5ng/mL) or converted, purified gDNA sample | 6.0 μL |
| Total | 20.0 μL |
Table 1: PCR mix.
For research use only. Not for use in diagnostic procedures. ©2023 MethylDetect. All rights reserved.
The PCR amplification and subsequent high-resolution melting were performed on the LightCycler® 480 Instrument II in LightCycler® 96-well plates using the setting Detection format » SYBR Green I/HRM dye».
| PCR program | Cycles | Temperature (°C) | Acquisition mode | Hold (sec) | Ramp rate (°C/sec) | Acquisitions (per °C) |
| Pre-incubation | 1 | 95 | None | 600 | 04.04 | – |
| Amplification | 50 |
95 BRCA1: 56 72 |
None
None
Single |
15
10
15 |
04.04
02.02
04.04 |
–
–
– |
| High-resolution melting | – | 95 60 95 |
None None Continuous |
15 60 – |
04.04 02.02 00.01 |
– – 50 |
Table 2: PCR and HRM program. The annealing temperature is specific to the assay and may vary between platforms.
The LightCycler® 480 Instrument II program for PCR amplification and the high-resolution melting are shown in Table 2. The listed annealing temperature is dependent on the assay. An annealing temperature of 56°C was used for EpiMelt BRCA1, 59°C for EpiMelt BRCA2, 50°C for EpiMelt HORMAD1, 59°C for EpiMelt MGMT, and 60°C for EpiMelt MLH1. No template controls were included for each assay, and all reactions were analyzed in triplicates. Data were analyzed using the LightCycler® 480 Software.
Results
The use of the EpiMelt Methylation Detection Assays and the EpiMelt Bisulfite Modification Standard Kit resulted in efficient bisulfite modification of all nine samples, followed by sensitive methylation detection. This is illustrated by the amplification curves (Figure 2A – E) and the relative signal difference (-d/dT) plots (Figure 2F – J) for each assay, and the Ct values for each sample and the EpiMelt Controls (Table 3).
The sample amplification curves were steep and reproducible for all EpiMelt Methylation Detection Assay replicates (Figure 2A-I). The high amplification signals and low Ct values (Table 3) suggest excellent template recovery and preservation of DNA integrity. In the relative difference plots, the peaks are uniform, and all three controls are clearly separable (Figure 2F-J). The melting profiles of the samples suggest complete bisulfite conversion, as they display uniform melting profiles that are easily comparable to the controls.
For research use only. Not for use in diagnostic procedures. ©2023 MethylDetect. All rights reserved.
Figure 2.
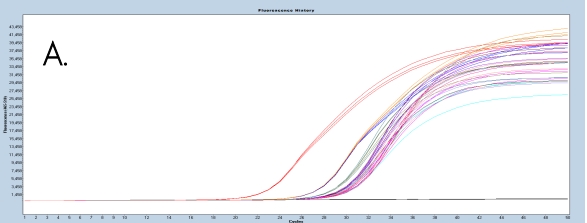
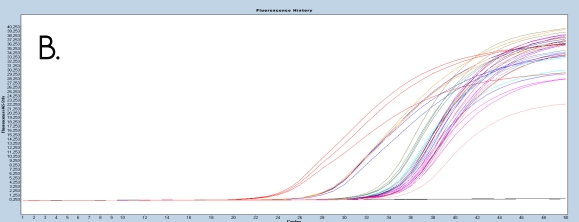
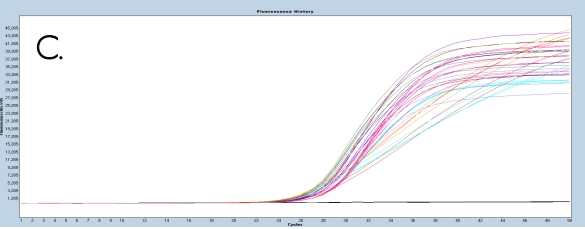
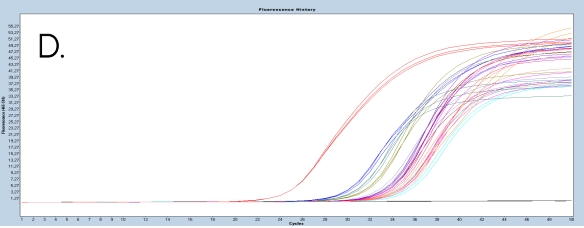
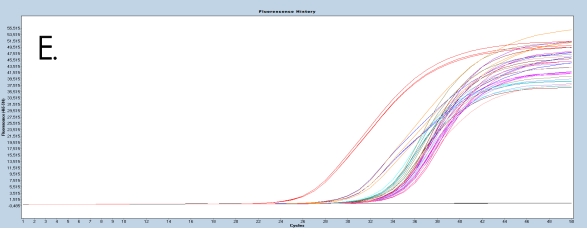
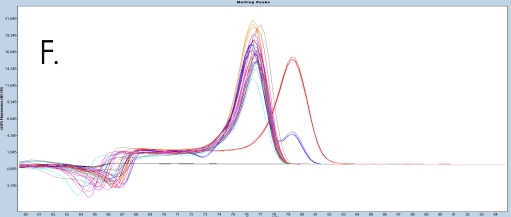
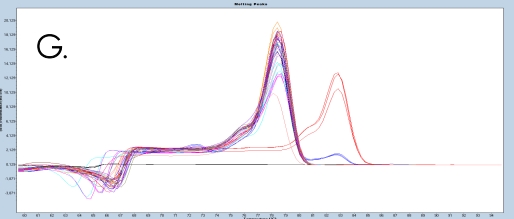
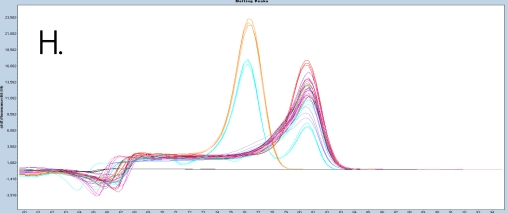
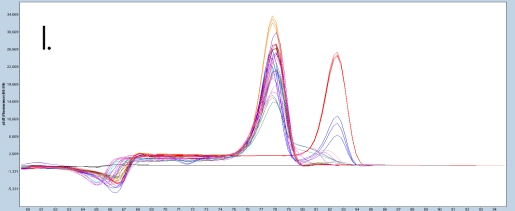
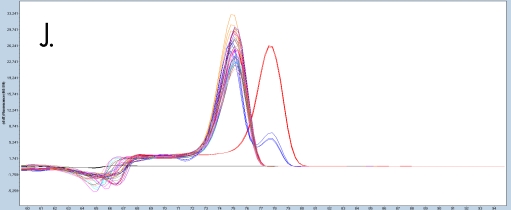

Figure 2: Amplification of nine FFPE DNA NSCLC samples and EpiMelt Controls using the EpiMelt BRCA1 Assay (A), EpiMelt BRCA2
Assay (B), EpiMelt HORMAD1 Assay (C), EpiMelt MGM1 Assay (D), and EpiMelt MLH1 Assay (E). Melting profiles of nine FFPE DNA.
For research use only. Not for use in diagnostic procedures. ©2023 MethylDetect. All rights reserved.
NSCLC samples and EpiMelt Control PCR products from EpiMelt BRCA1 Assay (F), EpiMelt BRCA2 Assay (G), EpiMelt HORMAD1
Assay (H), EpiMelt MGMT Assay (I), and EpiMelt MLH1 Assay (J).
| BRCA1 | BRCA2 | HORMAD1 | MLH1 | MGMT | |
| Methylation Positive Control | 22 | 24 | 27 | 25 | 24 |
| Assay Calibration Control | 26 | 27 | 26 | 31 | 29 |
| Methylation Negative Control | 26 | 27 | 26 | 31 | 34 |
| Sample 1 Low Quality | 30 | 34 | 28 | 33 | 35 |
| Sample 2 Low Quality | 30 | 34 | 28 | 34 | 33 |
| Sample 3 Low Quality | 30 | 34 | 28 | 34 | 34 |
| Sample 4 Medium Quality | 29 | 34 | 27 | 33 | 32 |
| Sample 5 Medium Quality | 30 | 35 | 28 | 34 | 34 |
| Sample 6 Medium Quality | 31 | 35 | 28 | 34 | 34 |
| Sample 7 High Quality | 29 | 34 | 27 | 32 | 30 |
| Sample 8 High Quality | 30 | 36 | 28 | 34 | 34 |
| Sample 9 High Quality | 30 | 35 | 28 | 34 | 34 |
Table 3: Ct values for the EpiMelt Controls and each gDNA sample for the five EpiMelt Methylation Detection Assays targeting BRCA1,
BRCA2, HORMAD1, MLH1, and MGMT.
Conclusion
In this application note, we demonstrate that the use of the EpiMelt Bisulfite Modification Standard Kit leads to efficient and complete bisulfite conversion of genomic DNA extracted from blood samples, with excellent template recovery. This is obtained with support from the EpiMelt Methylation Detection Assays, showing state-of-the-art methylation sensitivity.
Download the Application Note here:
References
1. Clark, S. J., Statham, A., Stirzaker, C., Molloy, P. L. & Frommer, M. DNA methylation: Bisulphite modification and analysis. Nature protocols 1,
2353-2364 (2006). https://doi.org/10.1038/nprot.2006.324
2. Nishiyama, A. & Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet 37, 1012-1027 (2021). https://doi.org/10.1016/j.tig.2021.05.002
3. Unnikrishnan, A. et al. The role of DNA methylation in epigenetics of aging. Pharmacol Ther 195, 172-185 (2019).
https://doi.org/10.1016/j.pharmthera.2018.11.001
4. Miura, F., Enomoto, Y., Dairiki, R. & Ito, T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids
Research 40, e136-e136 (2012). https://doi.org/10.1093/nar/gks454
5. Tanaka, K. & Okamoto, A. Degradation of DNA by bisulfite treatment. Bioorganic & Medicinal Chemistry Letters 17, 1912-1915 (2007).
https://doi.org/https://doi.org/10.1016/j.bmcl.2007.01.040
6. Yi, S., Long, F., Cheng, J. & Huang, D. An optimized rapid bisulfite conversion method with high recovery of cell-free DNA. BMC Molecular
Biology 18, 24 (2017). https://doi.org/10.1186/s12867-017-0101-4
7. Genereux, D. P., Johnson, W. C., Burden, A. F., Stöger, R. & Laird, C. D. Errors in the bisulfite conversion of DNA: modulating inappropriate- and
failed-conversion frequencies. Nucleic Acids Res 36, e150 (2008). https://doi.org/10.1093/nar/gkn691
8. Wojdacz, T. K., Hansen, L. L. & Dobrovic, A. A new approach to primer design for the control of PCR bias in methylation studies. BMC Res Notes
1, 54 (2008). https://doi.org/10.1186/1756-0500-1-54
9. Hussmann, D. & Hansen, L. L. Methylation-Sensitive High Resolution Melting (MS-HRM). Methods in molecular biology (Clifton, N.J.) 1708, 551-
571 (2018). https://doi.org/10.1007/978-1-4939-7481-8_28
10. Wojdacz, T. K. & Dobrovic, A. Melting curve assays for DNA methylation analysis. Methods in molecular biology (Clifton, N.J.) 507, 229-240
(2009). https://doi.org/10.1007/978-1-59745-522-0_17
